Our Research Projects
How important is mRNA localization in neurons?
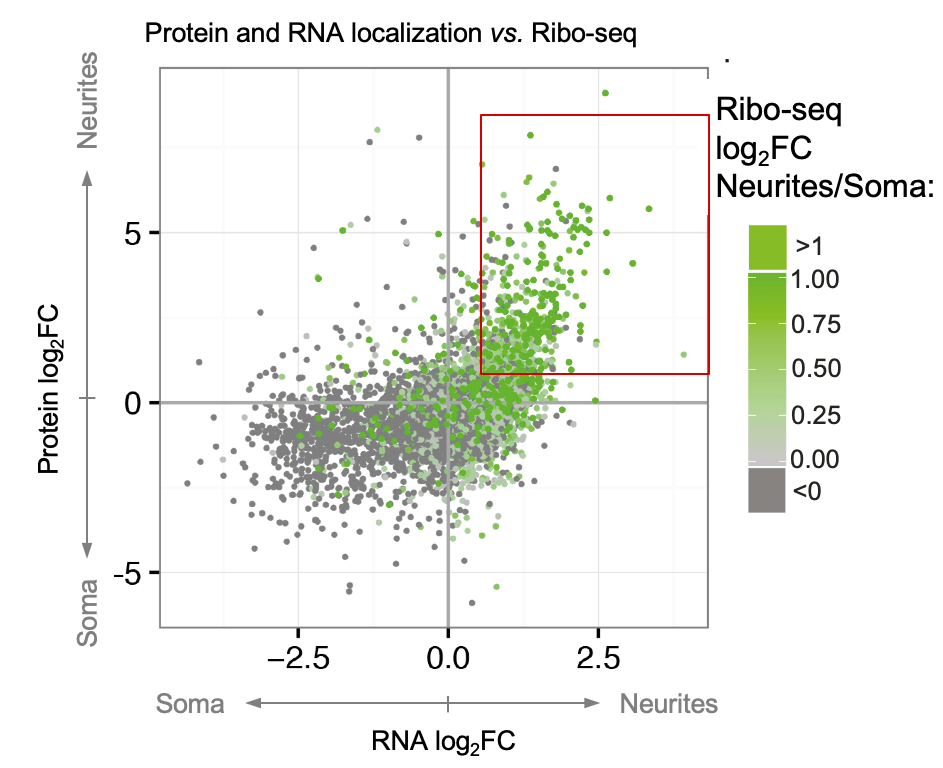
Cells adopt highly polarized shapes and form distinct subcellular compartments largely due to the localization of many proteins with local functions to specific areas. Protein localization can be achieved (1) by transporting proteins as parts of RNPs or vesicular organelles, (2) through mRNA localization and local translation, or (3) via local translation of equally distributed mRNAs. Specific examples of these mechanisms have been described in the literature. However, it has yet to be determined how much each of these mechanisms contributes to the overall asymmetry of protein distribution in neurons. To fill in this gap, we developed a neuron separation scheme in combination with mass spectrometry, RNA-seq, Ribo-seq, and bioinformatic analyses to identify proteins and RNAs that are differentially localized and translated between neurites and soma (spatial omics). This analysis identified hundreds of localized mRNAs and resulted in two key messages: (1) almost half of the neurite-enriched proteome is encoded by neurite-localized mRNAs, revealing mRNA localization as a key determinant of protein localization to neurites; (2) we generated a unique resource of the local neuronal transcriptome, proteome, and translatome and identified dozens of neurite-targeted non-coding RNAs and RNA-binding proteins with potential regulatory roles in neuronal polarity (Zappulo et al. Nature Comm 2017).
Which mRNAs localize in neurons?
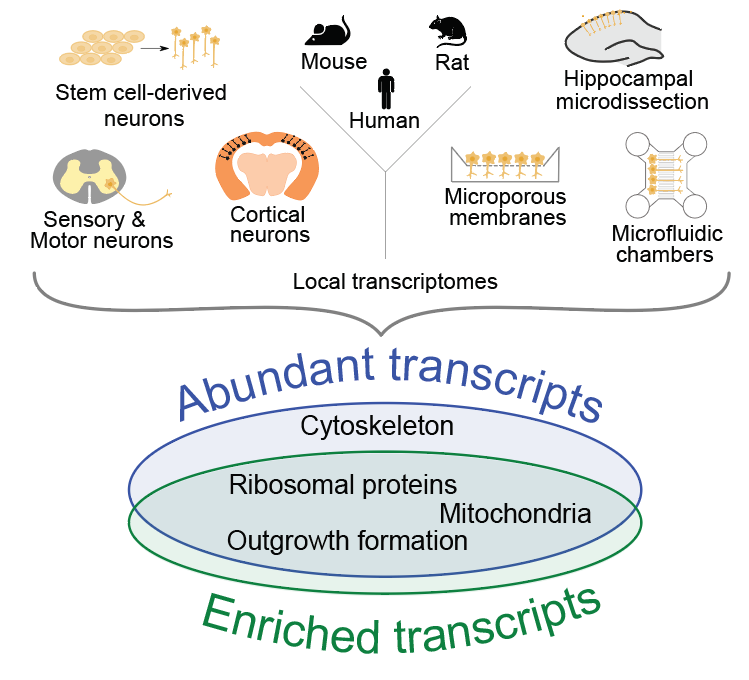
Recent high-throughput analyses have revealed that neurites contain hundreds to thousands of mRNAs, but an analysis comparing the transcriptomes derived from these studies has been lacking. We analyze 20 datasets pertaining to neuronal mRNA localization across species and neuronal types (von Kuegelgen and Chekulaeva WIRE RNA 2020). Our analysis has identified a conserved set of mRNAs that had robustly localized to neurites in a high number of studies. This set includes mRNAs encoding for ribosomal proteins and other components of the translation machinery, mitochondrial proteins, cytoskeletal components, and proteins associated with neurite formation. Our combinatorial analysis provides a unique resource for future hypothesis-driven research. To explore it in a user-friendly interface, check our lab resources.
What mediates mRNA localization in neurons?
Alternative splicing and polyadenylation
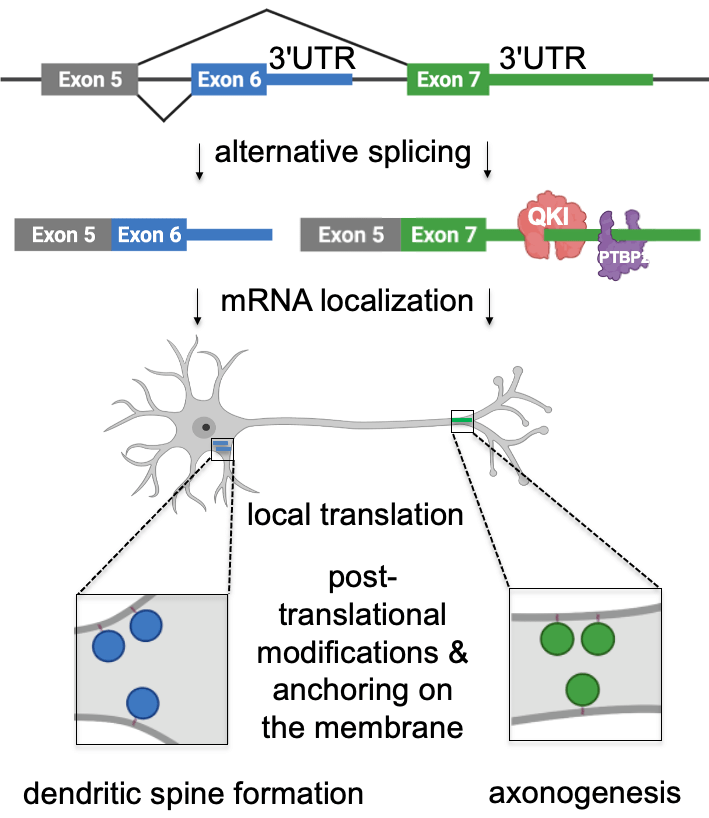
mRNA localization is mediated by sequences usually found in mRNA 3’UTRs. To identify these elements, we explored two directions. First, we investigated the processes that generate alternative 3’UTRs – alternative splicing and polyadenylation – and, therefore, have the potential to diversify mRNA localization patterns in neurons. For that, we performed mapping of alternative 3’UTRs in neurites and soma using 3′-end RNA-seq. Our analysis identified 593 genes with differentially localized 3’UTR isoforms. For example, we have shown that two isoforms of Cdc42 gene (cell division cycle 42) are differentially localized between neurites and soma due to their different 3’UTR sequences. Cdc42 is a small GTPase of the Rho family that shapes cellular morphology by controlling the actin cytoskeleton. Differential localization of its mRNA isoforms is essential in neurons because they produce two distinct protein isoforms with different functions in neuronal polarity. Thus, our work showed that the usage of alternative 3’UTRs serves as a novel mechanism mediating the localization of functionally distinct protein isoforms to different subcellular compartments (Ciolli Mattioli et al. NAR 2019).
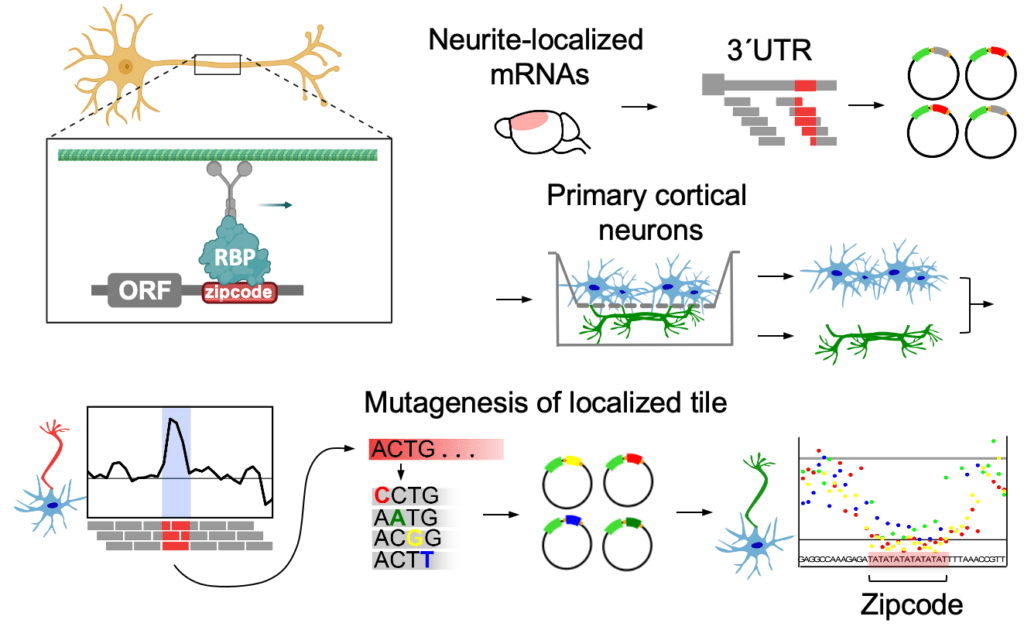
Cis-acting mRNA localization elements: zipcodes
How do these neuronal mRNAs find their way to their final destination? The localization is thought to be primarily mediated by cis-regulatory elements in the mRNA 3’UTRs – so-called “zipcodes”. Zipcodes are bound by specific RNA-binding proteins (RBPs) that link their targets to transport machinery or regulators of mRNA stability and direct their localization to the sites of function. While hundreds to thousands of mRNAs localize to neurites, only a few specific zipcodes and zipcode-bound RBPs have been described to date. To fill this gap, we developed a method to map and characterize neuronal zipcodes transcriptome-wide: neuronal zipcode identification protocol (N-zip). Our approach combines a massively parallel reporter assay with isolating neuronal subcellular compartments – soma and neurites. Using this method, we analyzed 99 neurite-localized transcripts. For one-third of them, we identified specific 3’UTR sequences localized to neurites of primary cortical neurons. These transcripts are associated with local neuronal structures and processes, such as synapse and actin cytoskeleton organization. Our further analysis identified the let-7 binding site and (AU)n motif as de novo zipcodes with functions in mouse primary cortical neurons (Mendonsa et al. Nature Neuroscience 2023 ).
How does RNA metabolism malfunction in neurodegeneration?
RNA mislocalization in ALS
It has been known for some time that the first signs of degeneration in many neurodegenerative diseases manifest in axons. These signs can be easily overlooked when analyses are performed on the whole neuron, as it has mostly been done up to now. Therefore, we are exploring the role of RNA localization and local translation in neurodegeneration. To this end, we have generated a biobank of hiPSC lines from the fibroblasts of amyotrophic lateral sclerosis (ALS) patients. ALS is a degenerative disease resulting in the progressive loss of motor neurons, eventually leading to paralysis and death. No curative treatment exists. We are applying spatial omics to understand how local RNA metabolism may malfunction in motor neurons differentiated from ALS patient-derived hiPSCs. This combined analysis will shed light on the fundamental links between ALS forms of different etiology and potentially identify new promising drug targets.
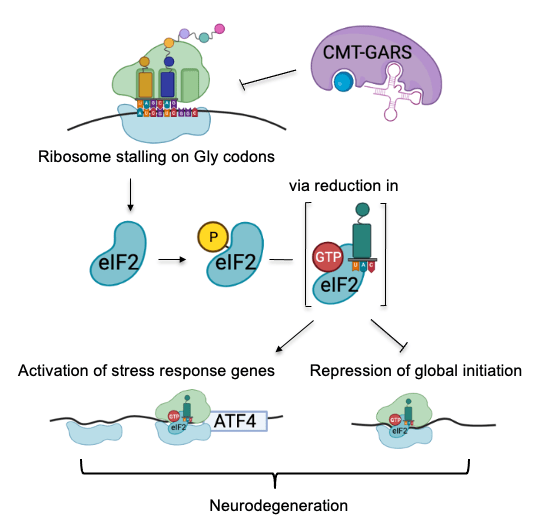
In Charcot–Marie–Tooth disease, mutant glycyl-tRNA synthetase represses translation via two mechanisms
We have also dissected the mechanism of translational repression in Charcot–Marie–Tooth (CMT) disease that causes the degeneration of motor and sensory neurons (Mendonsa et al. NAR 2021). CMT can be caused by mutations in aminoacyl-tRNA synthetases. While these mutations do not disrupt overall aminoacylation activity, they interfere with translation via an unknown mechanism. We used high-resolution ribosome profiling (link to methods) to dissect the mechanism of function of CMT mutant glycyl-tRNA synthetase (CMT-GARS). We found that CMT-GARS inhibits the first stage of elongation, the accommodation of glycyl-tRNA into the ribosomal A-site, causing ribosomes to pause at glycine codons. Moreover, ribosome pausing, in turn, activates a secondary repression mechanism at the level of translation initiation, the integrated stress response. Thus, the CMT-GARS mutant triggers translational repression via two interconnected mechanisms, affecting both elongation and initiation of translation.
How does SARS-CoV2 evade translational repression imposed by its own protein?
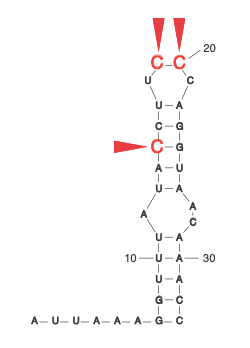
During the COVID-19 pandemic, we investigated how SARS-CoV-2 overcomes a conundrum faced by all viruses. To achieve its own replication and spread, it simultaneously depends on and subverts cellular mechanisms. At the early stage of infection, SARS-CoV-2 expresses the viral nonstructural protein 1 (NSP1), which inhibits host translation by blocking the mRNA entry tunnel on the ribosome. Viral mRNAs, however, find a way to overcome this blockade. Our analysis pinpointed specific residues within the stem-loop (SL1) in the viral leader that are required for viral evasion and thus might present promising drug targets. Our work, therefore, suggests a mechanism by which NSP1 inhibits the expression of host genes while enhancing that of viral RNA (Bujanic et al. RNA 2022).